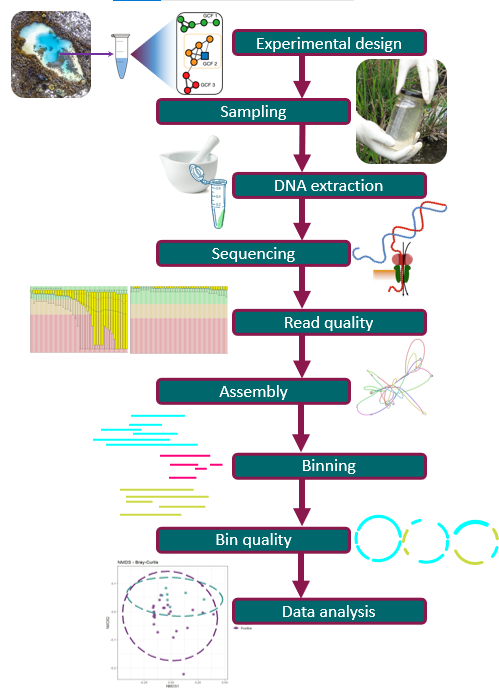
Starting a Metagenomics Project
Overview
Teaching: 15 min
Exercises: 15 minQuestions
How do you plan a metagenomics experiment?
How does a metagenomics project look like?
Objectives
Learn the differences between shotgun and metabarcoding (amplicon metagenomics) techniques.
Understand the importance of metadata.
Familiarize yourself with the Cuatro Ciénegas experiment.
Metagenomics
Metagenomes are collections of genomic sequences from various (micro)organisms that coexist in any given space. They are like snapshots that can give us information about the taxonomic and even metabolic or functional composition of the communities we decide to study. Thus, metagenomes are usually employed to investigate the ecology of defining characteristics of niches (* e.g.,*, the human gut or the ocean floor).
Since metagenomes are mixtures of sequences that belong to different species, a metagenomic workflow is designed to answer two questions:
- What species are represented in the sample?
- What are they capable of doing?
To find which species are present in a niche, we must do a taxonomic assignation of the obtained sequences. To find out their capabilities, we can look at the genes directly encoded in the metagenome or find the genes associated with the species that we found. In order to know which methodology we should use, it is essential to know what questions we want to answer.
Shotgun and amplicon
There are two paths to obtain information from a complex sample:
- Shotgun Metagenomics
- Metabarcoding.
Each is named after the sequencing methodology employed Moreover, have particular use cases with inherent advantages and disadvantages.
With Shotgun Metagenomics, we sequence random parts (ideally all of them) of the genomes present in a sample. We can search the origin of these pieces (i.e., their taxonomy) and also try to find to what part of the genome they correspond. Given enough pieces, it is possible to obtain complete individual genomes from a shotgun metagenome (MAGs), which could give us a bunch of information about the species in our study. MAGs assembly, however, requires a lot of genomic sequences from one organism. Since the sequencing is done at random, it needs a high depth of community sequencing to ensure that we obtain enough pieces of a given genome. Required depth gets exponentially challenging when our species of interest is not very abundant. It also requires that we have enough DNA to work with, which can be challenging to obtain in some instances. Finally, sequencing is expensive, and because of this, making technical and biological replicates can be prohibitively costly.
On the contrary, Metabarcoding tends to be cheaper, which makes it easier to duplicate and even triplicate them without taking a big financial hit. The lower cost is because Metabarcoding is the collection of small genomic fragments present in the community and amplified through PCR. Ideally, if the amplified region is present only once in every genome, we would not need to sequence the amplicon metagenome so thoroughly because one sequence is all we need to get the information about that genome, and by extension, about that species. On the other hand, if a genome in the community lacks the region targeted by the PCR primers, then no amount of sequencing can give us information about that genome. Conservation across species is why the most popular amplicon used for this methodology are 16S amplicons for Bacteria since every known bacterium has this particular region. Other regions can be chosen, but they are used for specific cases. However, even 16S amplicons are limited to, well, the 16S region, so amplicon metagenomes cannot directly tell us a lot about the metabolic functions found in each genome, although educated guesses can be made by knowing which genes are commonly found in every identified species.
On Metadata
Once we have chosen an adequate methodology for our study, we must take extensive notes on the origin of our samples and how we treated them. These notes constitute the metadata, or data about our data, and they are crucial to understanding and interpreting the results we will obtain later in our metagenomic analysis. Most of the time, the differences that we observe when comparing metagenomes can be correlated to the metadata, which is why we must devote a whole section of our experimental design to the metadata we expect to collect and record carefully.
Discussion #1: Choosing amplicon or shotgun sequencing?
Suppose you want to find the source of a nasty gut infection in people. Which type of sequencing methodology would you choose?
Which type of metadata would be helpful to record?Solution
For a first exploration, 16S is a better idea since you could detect known pathogens by knowing the taxons in the community. Nevertheless, if the disease is the consequence of a viral infection, the pathogen can only be discovered with shotgun metagenomics (that was the case of SARS-CoV 2). Also, metabarcoding does not provide insights into the genetic basis of the pathogenic phenotypes. Metadata will depend on the type of experiment. For this case, some helpful metadata could be sampling methodology, date, place (country, state, region, city, etc.), patient’s sex and age, the anatomical origin of the sample, symptoms, medical history, diet, lifestyle, and environment.
Cuatro Ciénegas
During this lesson, we will work with actual metagenomic information, so we should be familiarized with it. The metagenomes that we will use were collected in Cuatro Ciénegas, a region that has been extensively studied by Valeria Souza. Cuatro Ciénegas is an oasis in the Mexican desert whose environmental conditions are often linked to the ones present in ancient seas, due to a higher-than-average content of sulfur and magnesium but a lower concentrations of phosphorus and other nutrients. Because of these particular conditions, the Cuatro Ciénegas basin is a fascinating place to conduct a metagenomic study to learn more about the bacterial diversity that is capable to survive and thrive in that environment.
The particular metagenomic study that we are going to work with was collected in a study about the response of the Cuatro Cienegas’ bacterial community to nutrient enrichment. In this study, authors compared the differences between the microbial community in its natural, oligotrophic, phosphorus-deficient environment, a pond from the Cuatro Ciénegas Basin (CCB), and the same microbial community under a fertilization treatment. The comparison between bacterial communities showed that many genomic traits, such as mean bacterial genome size, GC content, total number of tRNA genes, total number of rRNA genes, and codon usage bias were significantly changed when the bacterial community underwent the treatment.
Exercise 1: Reviewing metadata
According to the results described for this CCB study.
What kind of sequencing method do you think they used, and why do you think so?
A) Metabarcoding
B) Shotgun metagenomics
C) Genomics of axenic culturesIn the table samples treatment information, what was the most critical piece of metadata that the authors took?
Solution
A) Metabarcoding. False. With this technique, usually, only one region of the genome is amplified.
B) Shotgun Metagenomics. True. Only shotgun metagenomics could have been used to investigate the total number of tRNA genes.
C) Genomics of axenic cultures. False. Information on the microbial community cannot be fully obtained with axenic cultures.The most crucial thing to know about our data is which community was and was not supplemented with fertilizers.
However, any differences in the technical parts of the study, such as the DNA extraction protocol, could have affected the results, so tracking those is also essential.
Exercise 2: Differentiate between IDs and sample names
Depending on the database, several IDs can be used for the same sample. Please open the document where the metadata information is stored. Here, inspect the IDs and find out which of them correspond to sample JP4110514WATERRESIZE
Solution
ERS1949771 is the SRA ID corresponding to JP4110514WATERRESIZE
Exercise 3: Discuss the importance of metadata
Which other information could you recommend to add in the metadata?
Solution
Metadata will depend on the type of the experiment, but some examples are the properties of the water before and after fertilization, sampling, and processing methodology, date and time, place (country, state, region, city, etc.).
Throughout the lesson, we will use the first four
characters of the File names (alias) to identify the data files
corresponding to a sample. We are going to use the first two sapmples for most of the lesson and the third one for one exercise at the end.
| SRA Accession | File name (alias) | Sample name in the lesson | Treatment |
|---|---|---|---|
| ERS1949784 | JC1ASEDIMENT120627 | JC1A | Control mesocosm |
| ERS1949801 | JP4DASH2120627WATERAMPRESIZED | JP4D | Fertilized pond |
| ERS1949771 | JP4110514WATERRESIZE | JP41 | Unenriched pond |
The results of this study, raw sequences, and metadata have been submitted to the NCBI Sequence Read Archive (SRA) and stored in the BioProject PRJEB22811.
Other metagenomic databases
The NCBI SRA is not the only repository for metagenomic information. There are other public metagenomic databases such as MG-RAST, MGnify, Marine Metagenomics Portal, Terrestrial Metagenome DB and the GM Repo.
Each database requires certain metadata linked with the data. As an example, when JP4D.fasta is uploaded to
mg-RAST the associated metadata looks like this:
| Column | Description |
|---|---|
| file_name | JP4D.fasta |
| investigation_type | metagenome |
| seq_meth | Illumina |
| project_description | This project is a teaching project and uses data from Okie et al. Elife 2020 |
| collection_date | 2012-06-27 |
| country | Mexico |
| feature | pond water |
| latitude | 26.8717055555556 |
| longitude | -102.14 |
| env_package | water |
| depth | 0.165 |
Key Points
Shotgun metagenomics can be used for taxonomic and functional studies.
Metabarcoding can be used for taxonomic studies.
Collecting metadata beforehand is fundamental for downstream analysis.
We will use data from a Cuatro Ciénegas project to learn about shotgun metagenomics.